Counting cells using YOLOv5
Contents
Counting cells using YOLOv5¶
YOLOv5: https://github.com/ultralytics/yolov5
Author of this notebook: Andre Telfer (andretelfer@cmail.carleton.ca)
pip install shapely
Requirement already satisfied: shapely in /home/andretelfer/anaconda3/envs/napari-env/lib/python3.9/site-packages (1.8.2)
Note: you may need to restart the kernel to use updated packages.
import matplotlib.pyplot as plt
import numpy as np
import shapely.geometry
import napari
import pandas as pd
from tqdm import tqdm
from pathlib import Path
from skimage.io import imread, imsave
What does our dataset look like?¶
Our dataset is just a folder containing .png images of cfos stains
DATA_DIR = Path("/home/andretelfer/shared/curated/brenna/cfos-examples/original")
plt.figure(figsize=(20,20))
sample_image = next(DATA_DIR.glob('*.png'))
image = imread(sample_image)
plt.imshow(image)
<matplotlib.image.AxesImage at 0x7f55958bcf40>
Create a new dataset by sampling sections of the original dataset¶
SIZE = 200 # size of new images in pixels
SAMPLES_PER_IMAGE = 5
def subsample_image(imagepath, samples):
image = imread(imagepath)
h,w,c = image.shape
locations = np.stack([
np.random.randint(0,h-SIZE,samples),
np.random.randint(0,w-SIZE,samples)
]).T
images = []
for (i,j) in locations:
new_image = np.zeros(shape=(SIZE,SIZE))
new_image = image[i:i+SIZE, j:j+SIZE]
images.append({
'image': new_image,
'x': j,
'y': i,
'path': imagepath
})
return images
sampled_images = []
for image in DATA_DIR.glob('*.png'):
sampled_images += subsample_image(image, SAMPLES_PER_IMAGE)
plt.imshow(sampled_images[5]['image'])
<matplotlib.image.AxesImage at 0x7f5594517c10>
Save the images to a new directory¶
OUTPUT_DIR = Path("/home/andretelfer/shared/curated/brenna/cfos-examples/subsamples")
! rm -rf {OUTPUT_DIR}
! mkdir -p {OUTPUT_DIR}
for item in sampled_images:
image = item['image']
fname = item['path'].parts[-1].split('.')[0]
output_file = f"{fname}_{item['x']}x_{item['y']}y.png"
imsave(OUTPUT_DIR / output_file, image)
ls {OUTPUT_DIR}
Rat2slide1sample3-L_305x_394y.png Rat2slide1sample4-R_100x_1311y.png
Rat2slide1sample3-L_510x_487y.png Rat2slide1sample4-R_1367x_1096y.png
Rat2slide1sample3-L_519x_748y.png Rat2slide1sample4-R_1522x_349y.png
Rat2slide1sample3-L_55x_1035y.png Rat2slide1sample4-R_1703x_387y.png
Rat2slide1sample3-L_624x_961y.png Rat2slide1sample4-R_824x_172y.png
Rat2slide1sample3-R_1386x_11y.png Rat2slide1sample5-L_258x_655y.png
Rat2slide1sample3-R_15x_1024y.png Rat2slide1sample5-L_496x_1318y.png
Rat2slide1sample3-R_1620x_1176y.png Rat2slide1sample5-L_693x_447y.png
Rat2slide1sample3-R_44x_328y.png Rat2slide1sample5-L_705x_749y.png
Rat2slide1sample3-R_582x_896y.png Rat2slide1sample5-L_79x_700y.png
Rat2slide1sample4-L_1376x_1171y.png Rat2slide1sample5-R_1320x_1213y.png
Rat2slide1sample4-L_1546x_250y.png Rat2slide1sample5-R_1474x_655y.png
Rat2slide1sample4-L_1550x_435y.png Rat2slide1sample5-R_1519x_298y.png
Rat2slide1sample4-L_217x_33y.png Rat2slide1sample5-R_476x_544y.png
Rat2slide1sample4-L_23x_960y.png Rat2slide1sample5-R_601x_334y.png
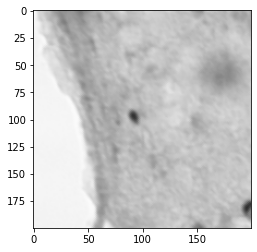
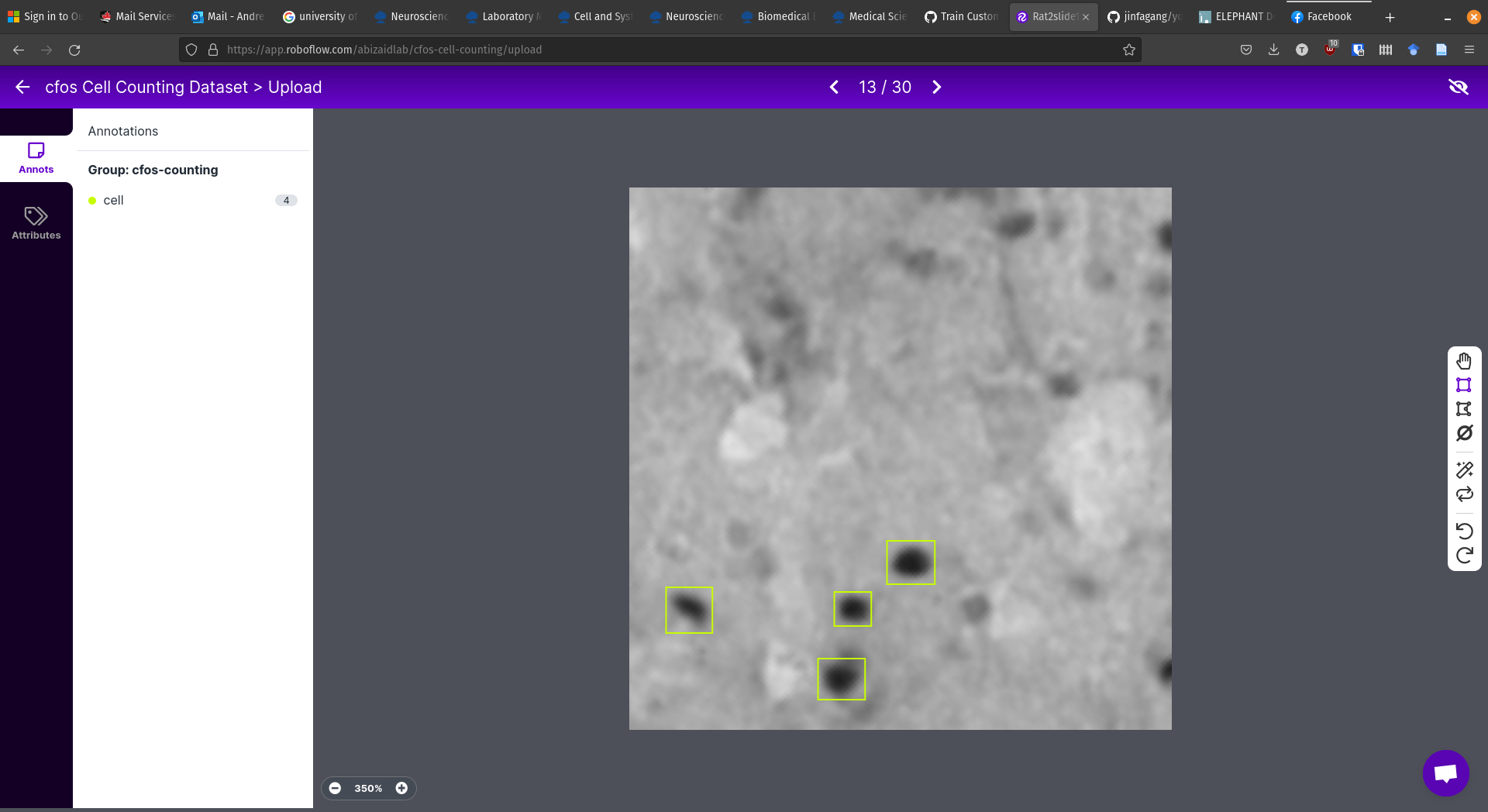
Labeling the Images¶
For this step, the YOLOv5 documentation recommended roboflow.
I created a free-tier account and started labelling.
I modified the tutorial by not including a scaling preprocessing step. I did this because the size of the cell matters and I wanted to preserve that information
there are some large splotches that are cell shaped, but are not cells
there are small speckles which are not cells
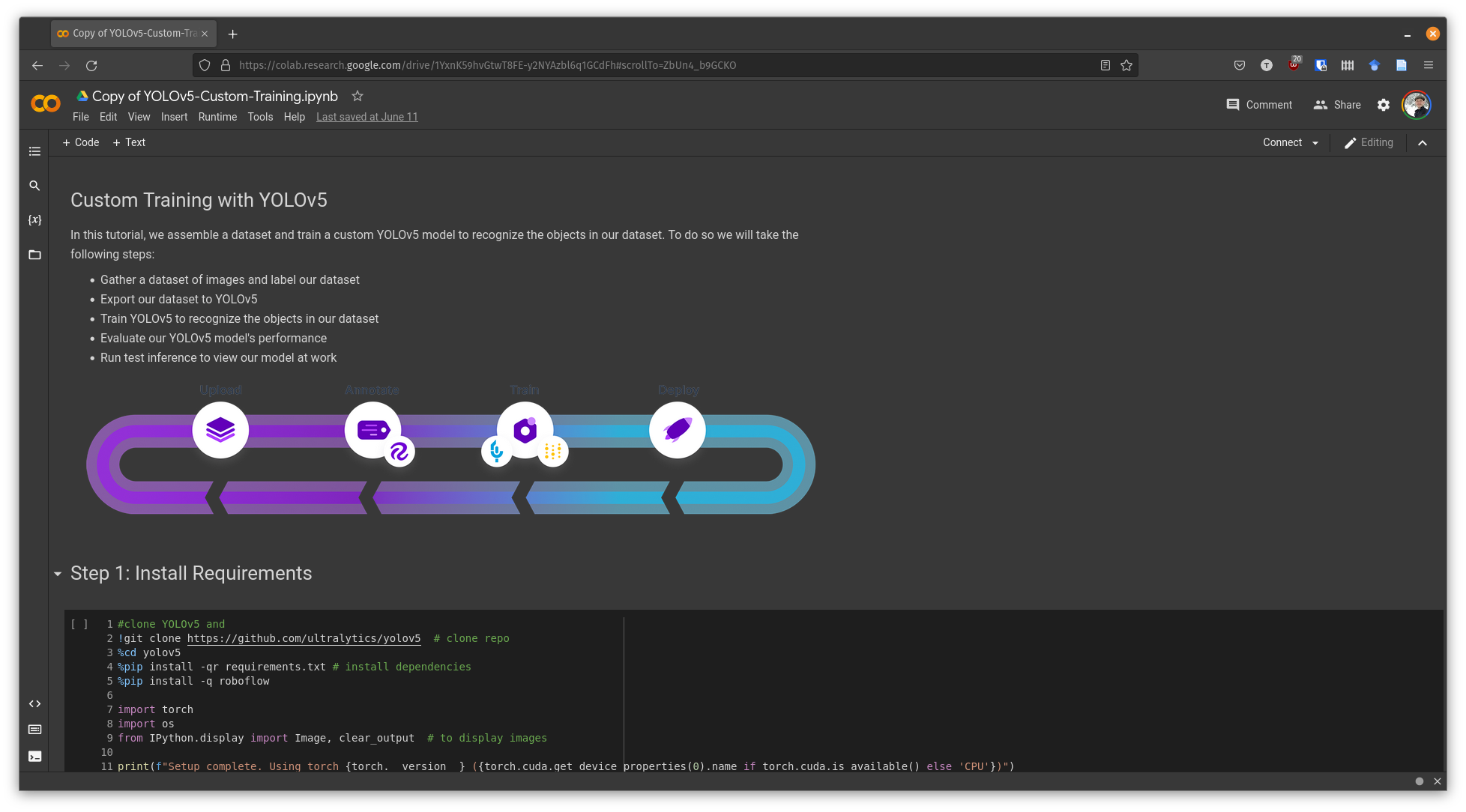
Training a Model¶
The YOLOv5 guide came with a Google Colab notebook that was easy to modify to my own examples (dataset, image sizes, etc)
Following the guide, I changed the dataset to the one we created in RoboFlow
In order to preserve information about cell size, for training I set the image size to be the same as the actual image size for the sampled training images. When running inference/detection, I used the full image size (e.g. ~2000px in my case).
Inference and Results¶
I uploaded the original images to google drive separately and modified the notebook to use them
The results were overall quite good, although I later found I should’ve labeled more of the lighter cells in the training data; so the model also misses the lighter cells.

Interacting with the results (correcting/viewing)¶
This will allow us to add/remove cells that were missed
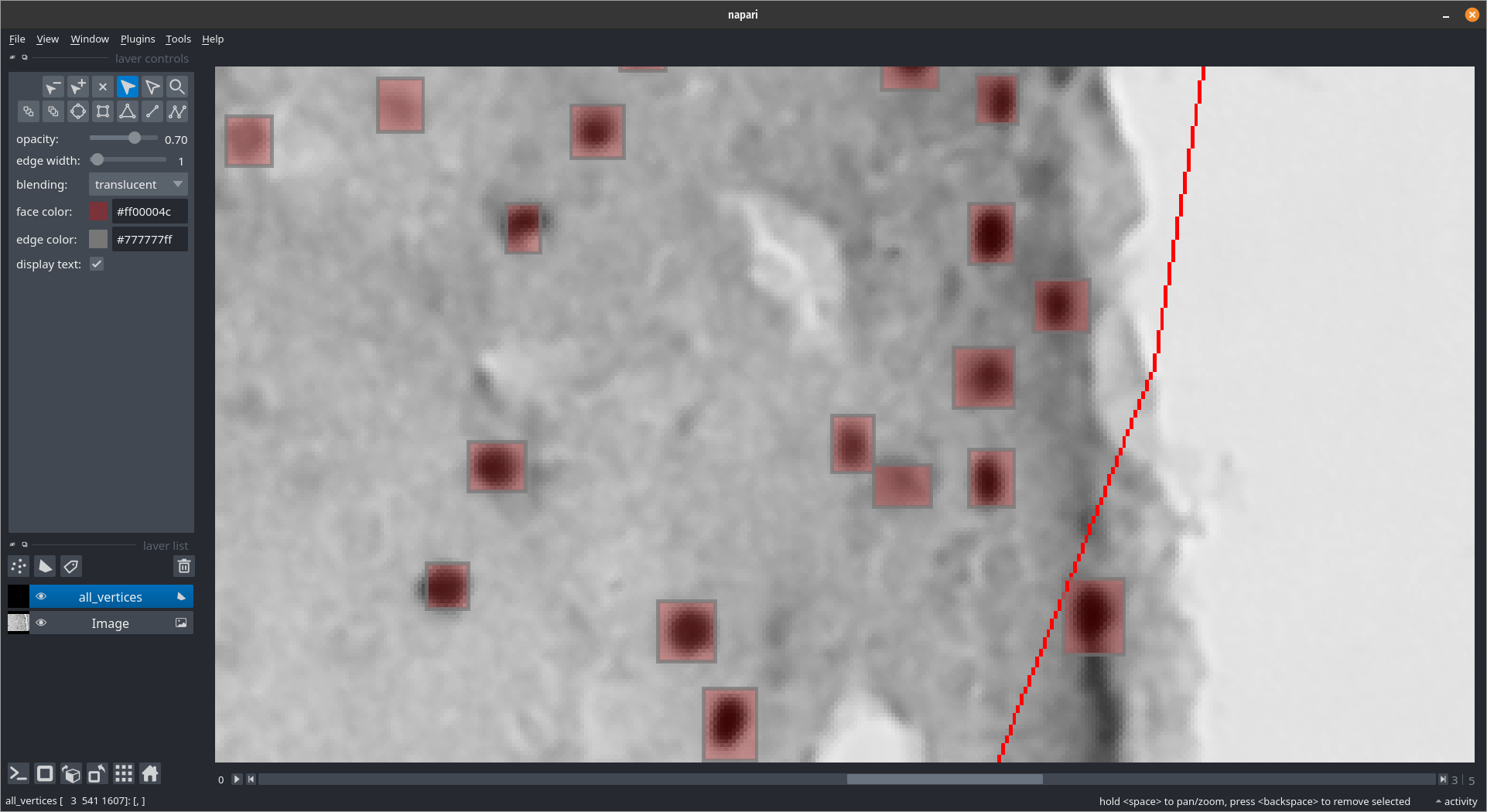
Loading the YOLO labels¶
all_vertices = []
for idx, p in enumerate(sorted(LABEL_DIR.glob("*.txt"))):
with open(p, 'r') as fp:
lines = fp.readlines()
# Turn the data into a dataframe
data = [l.strip().split(' ') for l in lines]
df = pd.DataFrame(data, columns=['0', 'x', 'y', 'w', 'h', 'c']).astype(float)
# Drop low confidence frames
df = df[df.c > 0.2]
# Scale by image size
fname, _ = p.parts[-1].split('.')
image = imread(DATA_DIR / f"{fname}.png")
h, w, c = image.shape
df.y *= h
df.h *= h
df.x *= w
df.w *= w
# Get x,y vertices for rectangle
i = np.ones(shape=df.shape[0])*idx
vertices = np.array([
[i, df.y-df.h/2, df.x-df.w/2],
[i, df.y+df.h/2, df.x-df.w/2],
[i, df.y+df.h/2, df.x+df.w/2],
[i, df.y-df.h/2, df.x+df.w/2]
]).transpose(2, 0, 1)
all_vertices.append(vertices)
all_vertices = np.concatenate(all_vertices)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Input In [8], in <cell line: 3>()
1 all_vertices = []
----> 3 for idx, p in enumerate(sorted(LABEL_DIR.glob("*.txt"))):
4 with open(p, 'r') as fp:
5 lines = fp.readlines()
NameError: name 'LABEL_DIR' is not defined
Viewing them with Napari¶
# The YOLOv5 labels
LABEL_DIR = Path("assets/yolov5-results/labels")
viewer = napari.Viewer()
# Add the images
images = np.array([imread(p) for p in sorted(DATA_DIR.glob("*.png"))])
image_layer = viewer.add_image(np.array(images))
# Add the yolov5 labels
shape_layer = viewer.add_shapes(all_vertices, face_color=[1., 0., 0., 0.3])
Getting cell counts¶
shape_layer.save('assets/cells.csv')
cells_df = pd.read_csv('assets/cells.csv')
cells_df.head(3)
| index | shape-type | vertex-index | axis-0 | axis-1 | axis-2 | |
|---|---|---|---|---|---|---|
| 0 | 0 | rectangle | 0 | 0.0 | 1041.999713 | 255.000033 |
| 1 | 0 | rectangle | 1 | 0.0 | 1054.999711 | 255.000033 |
| 2 | 0 | rectangle | 2 | 0.0 | 1054.999711 | 264.000031 |
cells_df = cells_df.rename(columns={'axis-0': 'image', 'axis-1': 'y', 'axis-2' : 'x', 'index': 'cell'})
cells_df.head(5)
| cell | shape-type | vertex-index | image | y | x | |
|---|---|---|---|---|---|---|
| 0 | 0 | rectangle | 0 | 0.0 | 1041.999713 | 255.000033 |
| 1 | 0 | rectangle | 1 | 0.0 | 1054.999711 | 255.000033 |
| 2 | 0 | rectangle | 2 | 0.0 | 1054.999711 | 264.000031 |
| 3 | 0 | rectangle | 3 | 0.0 | 1041.999713 | 264.000031 |
| 4 | 1 | rectangle | 0 | 0.0 | 17.000033 | 257.000059 |
Finally, we can get the cell counts for each image
cell_counts = cells_df.groupby('image').apply(lambda x: len(x.cell.unique()))
image
0.0 507
1.0 678
2.0 637
3.0 681
4.0 499
5.0 685
Name: cell_count, dtype: int64
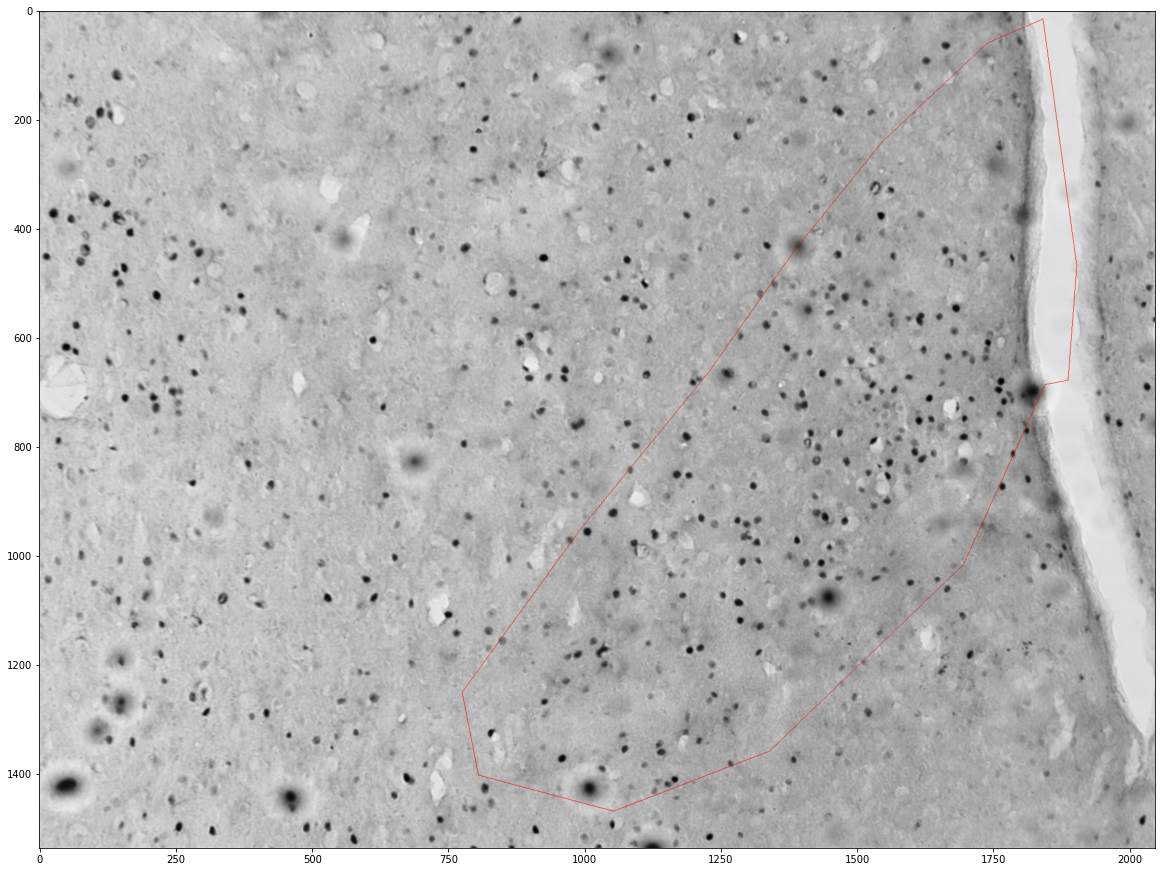
Getting cells in an area¶
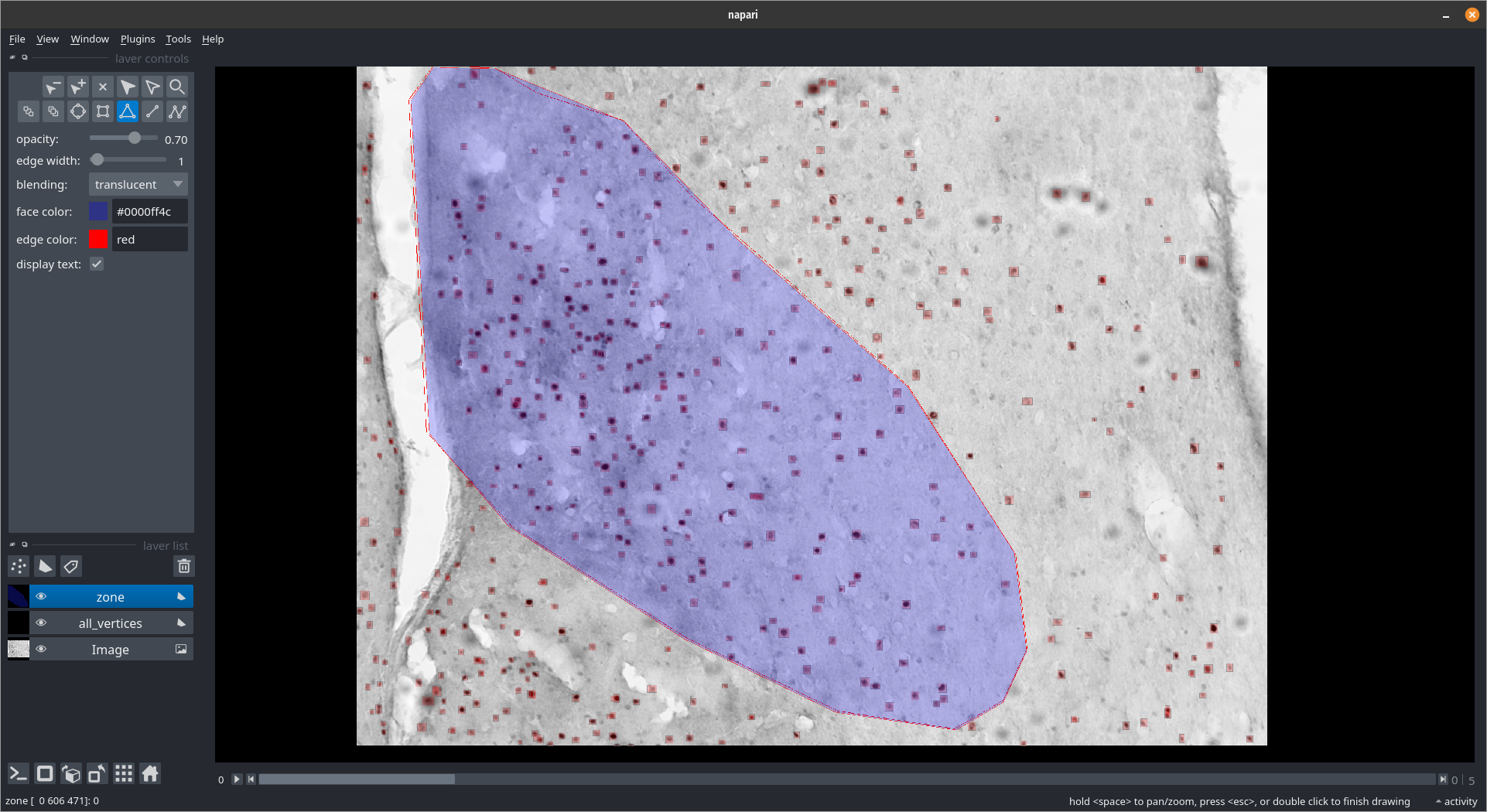
zone_layer = viewer.add_shapes(name='zone', ndim=3, edge_color='red', face_color=[0.,0.,1.,0.3])
zone_layer.save('assets/zones.csv')
zone_df = pd.read_csv('assets/zones.csv')
zone_df = zone_df.rename(columns={'axis-0': 'image', 'axis-1': 'y', 'axis-2' : 'x'})
cells_by_image = cells_df.groupby('image')
zones_by_image = zone_df.groupby('image')
for (idx, zone), (idx, cells) in zip(zones_by_image, cells_by_image):
zone = shapely.geometry.Polygon(zone[['x', 'y']].values)
plt.figure(figsize=(16,16))
plt.imshow(images[int(idx)])
x,y = zone.exterior.xy
plt.plot(x,y)
ax = plt.gca()
count = 0
for _, cell in tqdm(cells.groupby('cell')):
cell = shapely.geometry.Polygon(cell[['x', 'y']].values)
if zone.contains(cell):
count += 1
ax.add_patch(plt.Polygon(np.stack(cell.exterior.xy).T, color='red'))
plt.show()
print("Cell count", count)
print("Density", count / zone.area * 1e6)
print()
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 507/507 [00:00<00:00, 2086.06it/s]
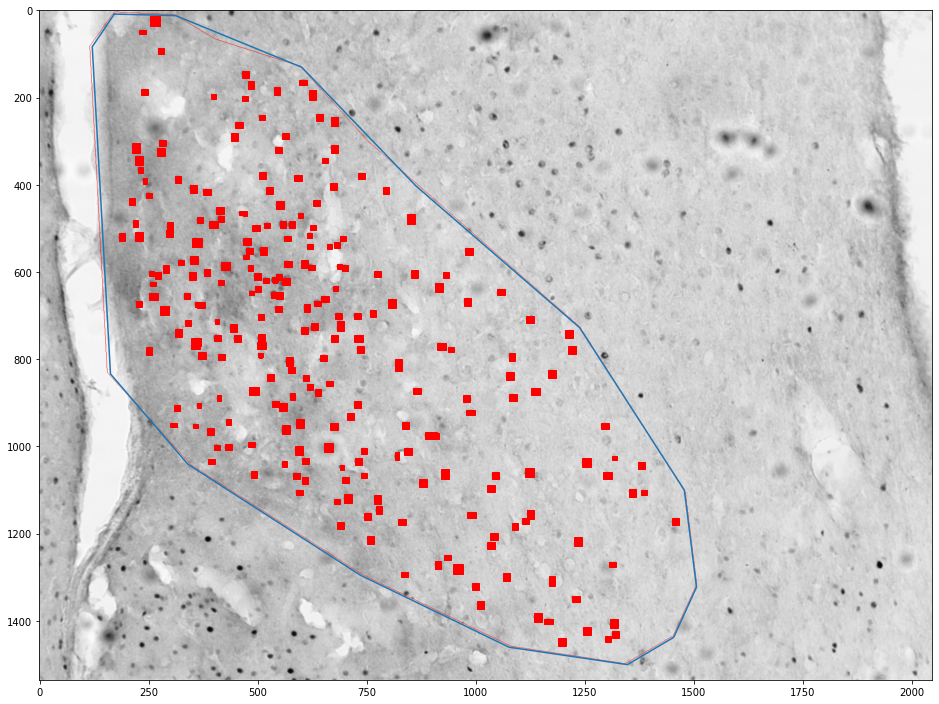
Cell count 253
Density 212.8925598329929
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 678/678 [00:00<00:00, 2406.08it/s]
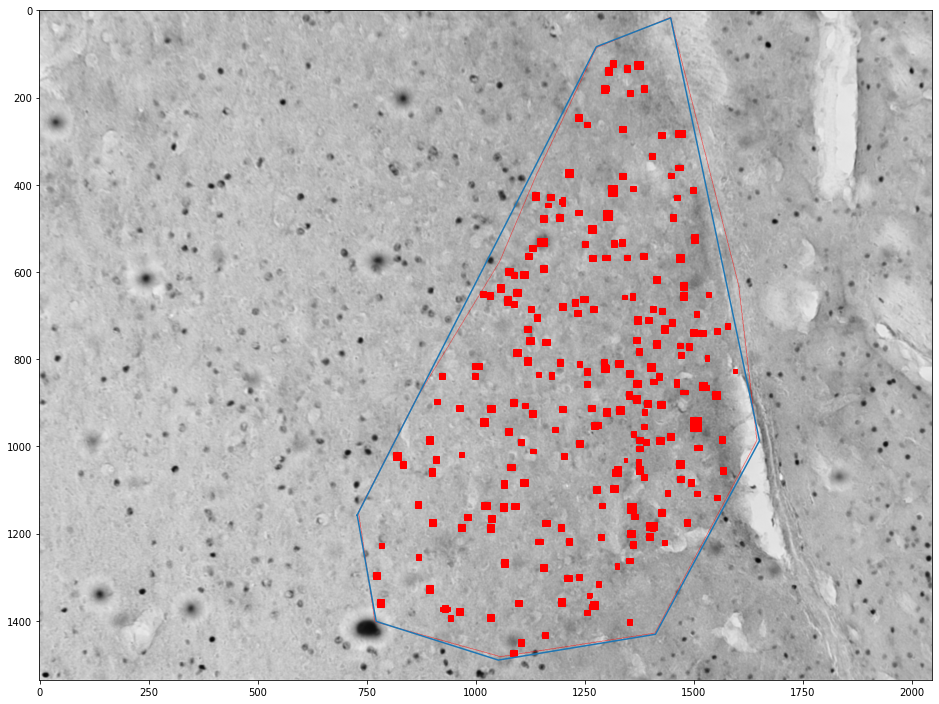
Cell count 237
Density 287.37546350839864
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 637/637 [00:00<00:00, 2235.87it/s]
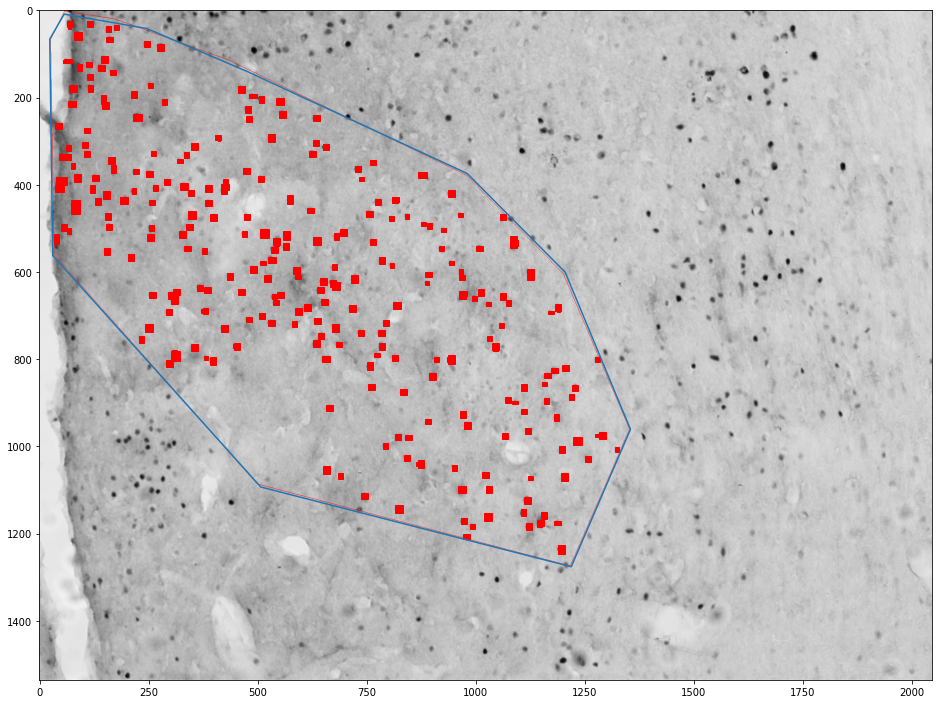
Cell count 280
Density 277.3213753614854
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 681/681 [00:00<00:00, 2398.25it/s]
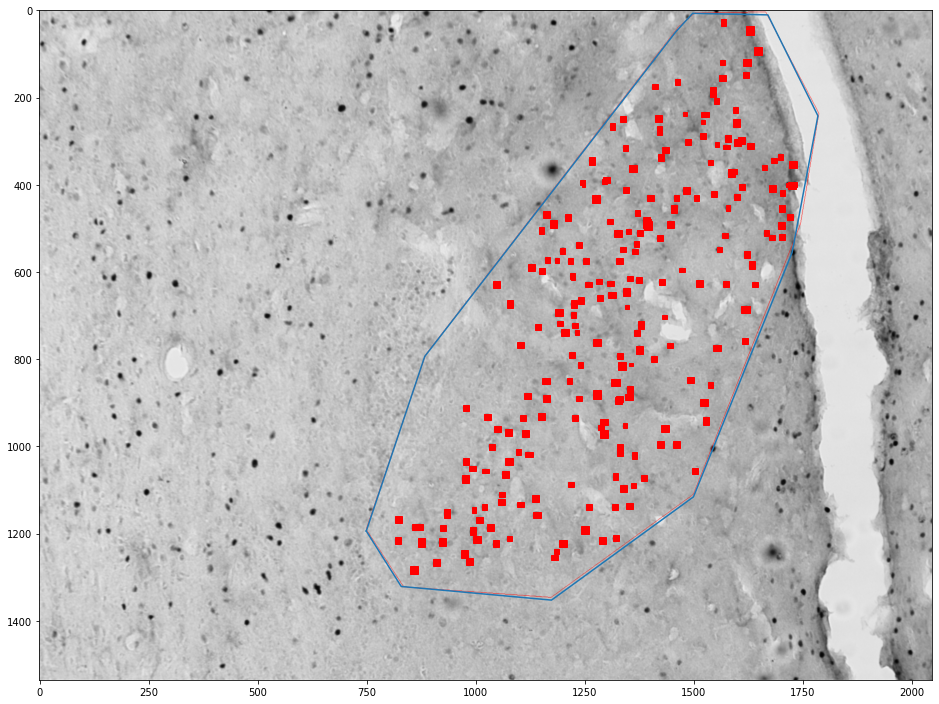
Cell count 231
Density 294.89183180541426
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 499/499 [00:00<00:00, 2362.33it/s]
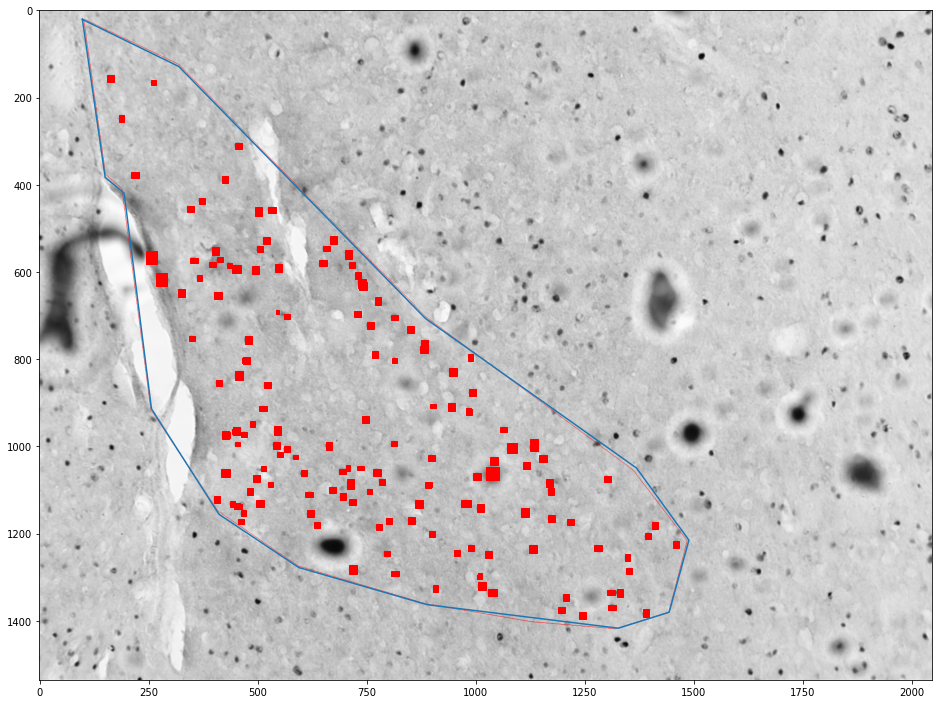
Cell count 155
Density 180.41495343681075
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 685/685 [00:00<00:00, 2353.12it/s]
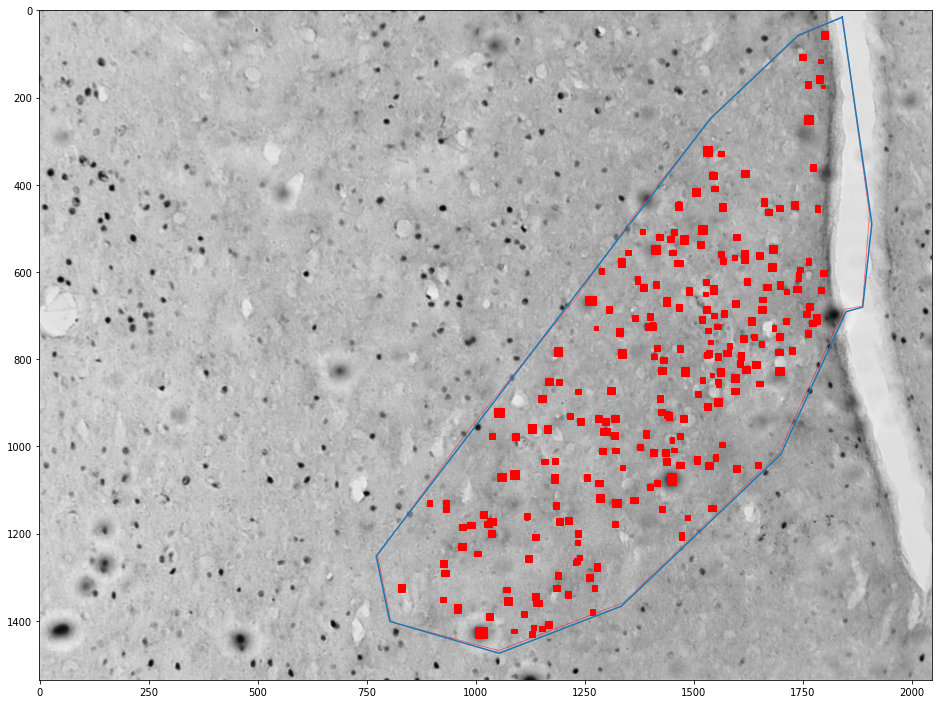
Cell count 238
Density 306.52820841199133
